
- SAP Community
- Groups
- Interest Groups
- Application Development
- Blog Posts
- K-Means Outlier Detection Algorithm ABAP Implement...
Application Development Blog Posts
Learn and share on deeper, cross technology development topics such as integration and connectivity, automation, cloud extensibility, developing at scale, and security.
Turn on suggestions
Auto-suggest helps you quickly narrow down your search results by suggesting possible matches as you type.
Showing results for
former_member26
Explorer
Options
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
02-03-2022
4:18 PM
Introduction:
Machine learning algorithms such as linear regression, logistic regression, decision tree are very popular topic in today’s market . This article focused on anomaly detection with k-means algorithm by using outliers.
K-means algorithm may be used on diverse of use case scenarios from image compression to system monitoring applications. Identifying anomaly in daily CPU resource utilization trends can be given as a sample use-case. Traditional system monitoring applications use fixed threshold values to identify a bottleneck over the system. But regarding today's requirements, it is not enough to monitor utilizations with fixed thresholds values. Monitoring system should to learn system resource utilization trends by using ML algorithms and create an alert in anomaly situation.
K-Means is a suitable solution for detecting anomalies in large data sets. Similarity analysis can be given as an example to detect anomalies in a dataset. In the example, two group of cars formed as two different clusters. Those clusters will have its own members in its immediate vicinity that is close similarity groups. If the element is close to the center of the cluster it becomes a group member. Elements far from the center, it identifies an anomaly situation for that cluster. If there is low similarity than those clusters, elements outside the clusters evaluates as an anomaly form.
K-means algorithm is basically based on Euclid calculation to measure distance of a tuple to centroid. This calculation repeating until distance values are being stable for each tuple. Please find k-Means visual sample on figure 1;
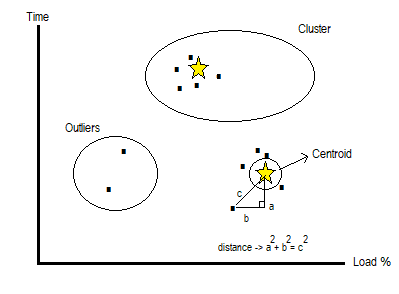
Figure 1
k-Means algorithm steps performed as below;
1- Assign each tuple to a randomly selected set
2- Calculate the centroid's distance from each cluster
3- Loop until there is no improvement or you reach the maxcount variable
4- Assign each tuple to the most appropriate cluster
5- Update cluster centroid
6- End loop
7- Return cluster informationHANA has its own k-means algorithm implementation in PAL library. This algorithm can be used easily by any kind of application that running a HANA system.
This article focused on k-Means outliers ABAP implementation without using HANA PAL library. Class has two main public functions. First one is “cluster” method. Second one is “outlier” method. In the cluster method, k-Means attribute values can be provided and assigned dynamically. It can have as many attributes as problem needs.
Dictionary objects:
Before coding, data types need to be defined at ABAP dictionary level.
Table types:
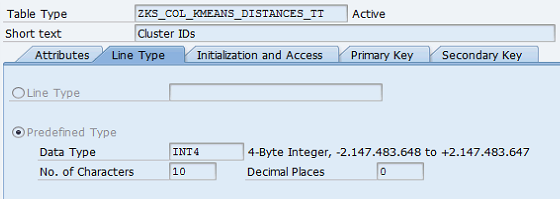
Figure 2

Figure 3

Figure 4

Figure 5

Figure 6
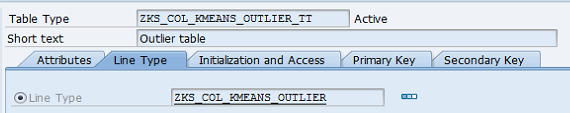
Figure 7

Figure 8
Structures:

Figure 9
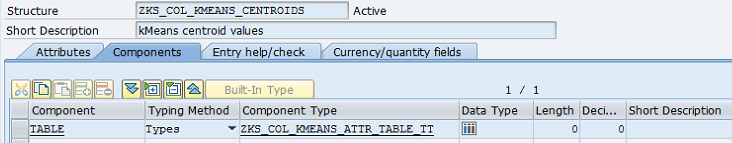
Figure 10

Figure 11

Figure 12

Figure 13
Test Program:
Dictionary objects figure 2, 3, 4, 5, 6, 7,8, 9, 10, 11, 12 ,13 should be created manually. Please find test program, below;
REPORT zkmeans_test.
TYPES: tt_atts TYPE STANDARD TABLE OF string WITH EMPTY KEY,
tt_rawdata TYPE TABLE OF zks_col_kmeans_rawdata WITH EMPTY KEY.
DATA: clustering TYPE REF TO zks_col_kmeans_clustering_tt,
outlier TYPE REF TO zks_col_kmeans_outlier_tt,
numattributes TYPE i,
numclusters TYPE i,
maxcount TYPE i,
lr_kmeans TYPE REF TO zcl_kmeans,
dataset TYPE tt_rawdata.
DATA(attributes) = VALUE tt_atts(
( `SampleAttribute1` )
).
dataset = VALUE #( ( table = VALUE #( ( value = 100 ) ) )
( table = VALUE #( ( value = 99 ) ) )
( table = VALUE #( ( value = 98 ) ) )
( table = VALUE #( ( value = 70 ) ) )
( table = VALUE #( ( value = 95 ) ) )
( table = VALUE #( ( value = 77 ) ) )
( table = VALUE #( ( value = 100 ) ) )
( table = VALUE #( ( value = 97 ) ) )
( table = VALUE #( ( value = 99 ) ) )
).
numattributes = lines( attributes ).
numclusters = 2.
maxcount = 30.
CREATE OBJECT lr_kmeans.
clustering = lr_kmeans->cluster(
rawdata = dataset
numclusters = numclusters
numattributes = numattributes
maxcount = maxcount ).
outlier = lr_kmeans->outlier(
rawdata = dataset
clustering = clustering
numclusters = numclusters
cluster = 0 ).In example above, only one attribute with a few data provided into the cluster method. Sample test program source code has only one centroid to make it easy to understand. Of course this value can be increased regarding the requirements. Max count value has been set to 30, in test program. But the higher this value, the better results it produces. In the example, in order to identifiy outlier values on cluster, outlier method has been called after cluster method. As soon as outlier values has been identified by the outlier method, those values can be dropped out from provided dataset to filter noisy values.
Class Implementation:
There are two public methods on ABAP ZCL_KMEANS class; cluster and outlier. Other methods are utility functions that help to execute k-Means algorithm. Please find method signatures and implementations by description;
|
|
|
|
|
|
|
|
|
|
|
Asymptotic Time Complexity:
Please find asymptotic time complexity analysis results of methods in K-Means class, below;
| Method | Value |
| CLUSTER | n^2 |
| INITCLUSTERING | n |
| ALLOCATECENTROIDS | n^2 |
| ALLOCATEMEANS | n^2 |
| UPDATEMEANS | n^2 |
| UPDATECENTROIDS | n |
| ASSIGN | n^2 |
| OUTLIER | n^2 |
| DISTANCE | n |
| MININDEX | n |
| COMPUTECENTROID | n^2 |
Summary:
This is an ABAP implementation of k-Means algorithm. Of course this code can be written more optimized way. Or may add and remove or merge with some other functionalities regarding problem needs.
Please find transport request files under Github. Import transport request into your sandbox or development system.
Reference:
Data Clustering - Detecting Abnormal Data Using k-Means Clustering
About Author:
Please find my personal bio orkun.gedik4#about;
I have been working at KoçSistem as a SAP basis technology executive team leader and principal SAP technical architect, since 2013. With my 26 years experience on ABAP development and 21 years experience on SAP basis fields, during my consulting engagements I worked on various technologies such as cloud, robotic programming, ML, mobile application development, application server programming and many fields and platforms. Currently studying at Gazi University MSc. Computer Science department, since 2021.
SAP Community Network topic leader @
- 2011/2012 - Databases and OS Platforms
- 2012/2013 - SAP On Oracle
- SAP Managed Tags:
- Machine Learning,
- ABAP Development
15 Comments
You must be a registered user to add a comment. If you've already registered, sign in. Otherwise, register and sign in.
Labels in this area
-
A Dynamic Memory Allocation Tool
1 -
ABAP
9 -
abap cds
1 -
ABAP CDS Views
14 -
ABAP class
1 -
ABAP Cloud
1 -
ABAP Development
5 -
ABAP in Eclipse
2 -
ABAP Keyword Documentation
2 -
ABAP OOABAP
2 -
ABAP Programming
1 -
abap technical
1 -
ABAP test cockpit
7 -
ABAP test cokpit
1 -
ADT
1 -
Advanced Event Mesh
1 -
AEM
1 -
AI
1 -
API and Integration
1 -
APIs
9 -
APIs ABAP
1 -
App Dev and Integration
1 -
Application Development
2 -
application job
1 -
archivelinks
1 -
Automation
4 -
B2B Integration
1 -
BTP
1 -
CAP
1 -
CAPM
1 -
Career Development
3 -
CL_GUI_FRONTEND_SERVICES
1 -
CL_SALV_TABLE
1 -
Cloud Extensibility
8 -
Cloud Native
7 -
Cloud Platform Integration
1 -
CloudEvents
2 -
CMIS
1 -
Connection
1 -
container
1 -
Customer Portal
1 -
Debugging
2 -
Developer extensibility
1 -
Developing at Scale
3 -
DMS
1 -
dynamic logpoints
1 -
Dynpro
1 -
Dynpro Width
1 -
Eclipse ADT ABAP Development Tools
1 -
EDA
1 -
Event Mesh
1 -
Expert
1 -
Field Symbols in ABAP
1 -
Fiori
1 -
Fiori App Extension
1 -
Forms & Templates
1 -
General
1 -
Getting Started
1 -
IBM watsonx
2 -
Integration & Connectivity
10 -
Introduction
1 -
JavaScripts used by Adobe Forms
1 -
joule
1 -
NodeJS
1 -
ODATA
3 -
OOABAP
3 -
Outbound queue
1 -
ProCustomer
1 -
Product Updates
1 -
Programming Models
14 -
Restful webservices Using POST MAN
1 -
RFC
1 -
RFFOEDI1
1 -
SAP BAS
1 -
SAP BTP
1 -
SAP Build
1 -
SAP Build apps
1 -
SAP Build CodeJam
1 -
SAP CodeTalk
1 -
SAP Odata
2 -
SAP SEGW
1 -
SAP UI5
1 -
SAP UI5 Custom Library
1 -
SAPEnhancements
1 -
SapMachine
1 -
security
3 -
SM30
1 -
Table Maintenance Generator
1 -
text editor
1 -
Tools
18 -
User Experience
6 -
Width
1
Top kudoed authors
| User | Count |
|---|---|
| 4 | |
| 4 | |
| 3 | |
| 3 | |
| 2 | |
| 2 | |
| 1 | |
| 1 | |
| 1 | |
| 1 |